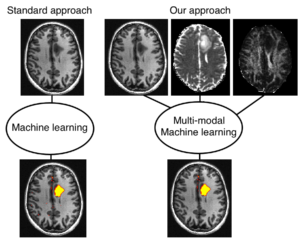
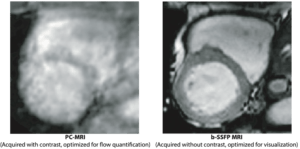
Innovationsprojekt
AIDAs utvecklingsprojekt tar fram AI-baserade beslutsstöd och drivs av forskargrupper inom industri och akademi från hela landet i samarbete med vårdgivare. Nedan finns sammanfattningar av AIDAs projekt.
AI-based prediction of magnetic resonance elastography
Rodrigo Moreno
Kungliga Tekniska Högskolan
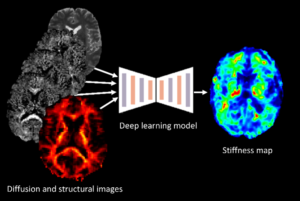 The project centers on developing AI-based tools that can estimate the mechanical properties of brain tissue, such as stiffness, by utilizing widely available single-shell diffusion MRI (dMRI) data. The endeavor seeks to provide a more accessible alternative to Magnetic Resonance Elastography (MRE), which is limited by the need for specialized equipment.
The project centers on developing AI-based tools that can estimate the mechanical properties of brain tissue, such as stiffness, by utilizing widely available single-shell diffusion MRI (dMRI) data. The endeavor seeks to provide a more accessible alternative to Magnetic Resonance Elastography (MRE), which is limited by the need for specialized equipment.
The first part of the project involves creating synthetic multi-shell dMRI data from single-shell scans using state-of-the-art generative AI techniques. This approach simulates the necessary data for mechanical property estimation without MRE hardware. In the second part, the focus shifts to predicting brain tissue stiffness. Here, deep learning models are trained in a supervised fashion using both synthetic dMRI data and actual tissue stiffness measurements.
The final part of the project applies the developed AI tools to the diagnosis and characterization of Parkinson’s Disease. By integrating various imaging modalities and employing models that can handle incomplete data, this stage aims to enhance the predictive diagnosis of Parkinson’s Disease and potentially other neurological conditions. The successful implementation of this project could transform the neuroimaging approach in clinical practice, making it more feasible for hospitals to assess brain tissue mechanics for diagnosis and treatment planning.
Federated learning for efficient radiotherapy treatment planning
Anders Eklund
Linköping University
Radiation therapy treatment planning can be time consuming, as it requires manual segmentation of the tumor and several organs at risk. In medical imaging, it has been demonstrated in many applications that deep learning segmentation models, such as the U-Net, can perform automatic segmentation in a few seconds (which can save time in the clinical workflow). However, deep learning models with millions of parameters require large, annotated datasets for supervised training, and the datasets need to be diverse in order for the models to generate good predictions for as many patients as possible. It is costly and time consuming to create large datasets, especially if the datasets should contain images and manual segmentations from several different hospitals. Many hospitals and researchers are reluctant to share their sensitive data, as it may violate GDPR or other legal or ethical regulations. The purpose of this project is therefore to demonstrate that it is possible to train segmentation models for radiation therapy treatment planning through federated learning, where a model can be trained without sending any image data between different hospitals. The future patient benefit is saved time in the clinical workflow, which can reduce waiting time.
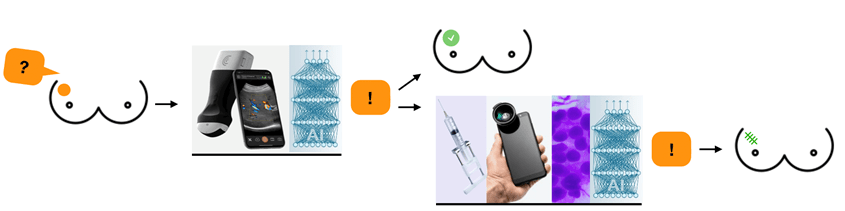
Breast Cancer Diagnostics in Low-Resource Settings Using Point-of-Care Ultrasound and Deep Learning
Despite breast cancer being the most common cancer globally, women in low-and middle-income countries have limited access to health care leading to late-stage diagnosis and poor survival rates. To address this shortness, we aim to develop an accessible breast diagnostic tool consisting of a point-of-care ultrasound (POCUS) device paired with a user-friendly, smartphone-based deep learning algorithm. We have developed an algorithm for breast cancer detection in POCUS images, and in this project we aim to continue develop the solution by (1) improvement of our POCUS vendor neutral algorithm, (2) develop an algorithm determining adequate image quality at acquisition and integrating uncertainty assessment in algorithm results, (3) develop a diagnostic prototype app with real time ultrasound images, algorithms, and user interface (4) perform a clinical study at the University Hospital in Skåne and a clinical feasibility study in Kenya. The overall aim of this project is to develop a diagnostic solution ready to be used in a clinical setting for a timely diagnosis of breast cancer in low-resource settings.
AI for complete pre-processed CMR in clinic
A medium-size hospital performs over 2000 exams using cardiac magnetic resonance (CMR) imaging per year. Given the large amount of imaging data generated for each examination, it is currently a major challenge for staff to find time to interpret the images and write reports. In this project, we will develop two AI-based analysis algorithms for fully automatic flow and infarct quantification. At the end of the project, when combined with existing software platform and AI-algorithms, this will allow for a complete pipeline where CMR images can be sent directly from the MR scanner, be automatically loaded into a software where a full analysis is performed by state-of-the-art AI algorithms. All the user needs to do is to review segmentations, correct if needed, and formulate the written patient report. We estimate that the time saved per patient will be approximately 10 minutes. For a medium-size hospital, approximately 2 months of a full-time position can be saved per year – time and resources better spent on patients.
Accurate, fast, and automatic assessment of whole-heart anatomy from 3D CT for early disease detection and diagnosis
Tino Ebbers
Linköping University
Coronary CT is frequently used for the diagnosis of chronic coronary syndromes. The acquired data contains valuable geometrical information about the heart including the volume and shape of the left atrium, which is a predictor of atrial fibrillation. However, this information about cardiac shape from CT is seldom used and the same information is difficult to measure accurately with other techniques. Fully automatic segmentations of the heart tend to fail in disease populations, making it difficult to analyze shape information from cardiac CT data at a large scale. This project aims to utilize 3T CT imaging of the heart from a large Swedish national cohort to improve current state-of-the-art deep learning methods for cardiac segmentation. The AI segmentation tool will be compared to established methods for left atrial shape and volume measurement and calibrated to accurately quantify prediction uncertainty across healthy individuals and patients. Using the AI-generated information about left atrial shape in a large population, this project will explore the role of early cardiac anatomical changes in disease detection and progression.
Development of an automated pipeline for the neuroradiological diagnosis of iNPH using Deep Learning
Johan Wikström
Dimitris Toumpanakis
Uppsala University
Idiopathic Normal Pressure Hydrocephalus (iNPH) is a form of communicating hydrocephalus accompanied often by the clinical triad of gait disturbance, cognitive impairment and urinary incontinence. The prevalence of iNPH is 3.7% among individuals 65 years and older with further increasing prevalence at higher ages. The condition is however believed to be under-diagnosed and under-treated and the diagnosis will be of increasing importance with increasing proportion of elderly subjects in the population. Early diagnosis is of paramount importance since iNPH is the only potentially reversible cause of dementia. If the diagnosis is made at the early stages of the disease, the patient can be treated via surgical placement of a VP-shunt in the brain with significant improvement of symptoms.
This project aims to develop an ensemble of neural networks that takes multiparametric input (various classical and volumetric neuroimaging biomarkers together with direct Magnetic Resonance Imaging input) and predicts the radiological probability for a patient having iNPH and for positive response after surgical treatment. The final goal is to have a clinically useful decision support tool for earlier, faster and more confident iNPH diagnosis and treatment.
Advancing Lumbar Spinal Stenosis Surgical Treatment Decision and Prognostic Outcomes through Imaging and AI
Christian Waldenberg
Sahlgrenska University Hospital
This project aims to improve the understanding and management of Lumbar Spinal Stenosis (LSS) by using AI-based MRI techniques to enhance surgical decision-making and patient outcomes. LSS, characterized by the narrowing of the lumbar spinal canal, often leads to lower back pain due to nerve compression. Traditional surgical approaches show mixed results due to factors like patient selection and individual risks, highlighting the need for advanced analysis and prognostic tools.
Utilizing advanced AI techniques like machine learning and texture analysis, the project aims to provide a detailed examination of spinal tissues, identifying abnormalities beyond the scope of traditional visual inspection. This analysis is expected to directly correlate with patient outcomes, including pain and treatment efficacy. The research is based on a comprehensive intervention study involving over 400 individuals across 17 centers. The focus is on understanding the effects of LSS on adjacent spinal tissues and developing MRI-based prognostic tools to predict surgical success, thereby refining treatment approaches and enhancing patient recovery.
Detect and monitor longitudinal domain shift for deep learning in digital pathology
Caroline Bivik Stadler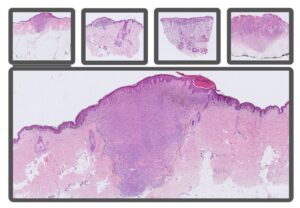
Linköping University
It is of utmost importance for AI models in healthcare to consistently deliver reliable results over time. One potential challenge faced by AI diagnostic systems is domain shift – the difference of data distribution between training and test datasets, often leading to degradation in AI model performance. The focus of this study is to investigate the impact of longitudinal domain shift on AI algorithms used in histopathology applications. Minor changes in the data preparation pipeline, such as staining protocol variations, software updates and new equipment, may influence model performance and resilience of an AI model over an extended period. To address this issue, we will study longitudinal shift within the context of tumor classification using an extensive histopathological skin dataset. The data will be annotated, and an AI model will be trained to classify lesions into four diagnoses: benign, malignant melanoma, basal cell carcinoma and squamous cell carcinoma. Thereafter, we will evaluate the model’s performance on datasets collected from the following years to determine when this model begins to exhibit signs of domain shift effects. Finally, we plan to develop an unsupervised method for detecting performance changes, enabling proactive interventions to maintain model efficacy deployed in a real clinical context and ensure patient safety.
Prediction of Treatment Outcome from PET-CT Imaging Data in Lymphoma
Joel Kullberg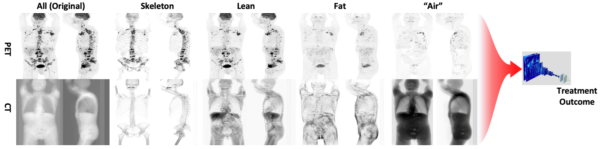
Uppsala University
The aim of this project is to develop and evaluate an automated methodology for enhanced predictions of treatment outcome from PET-CT images in patients with lymphoma. PET-CT imaging is widely used in diagnosis and treatment follow up in lymphoma and other cancer forms and contains very detailed information from each patient examination. Today, due to the lack of efficient tools, the image data is only sub optimally analysed using visual and semi-quantitative assessments, sometimes of a subset of the cancer lesion burden only. This project will optimize and evaluate an approach that attempts efficient analysis of all the very detailed information available in the PET-CT imaging data in the predictions. The described project will primarily use PET-CT imaging data collected in the U-CAN project (www.u-can.uu.se). Predicting who will benefit from a certain therapy is an important part of personalized medicine. For lymphoma where many treatments are available, and many are under development, good tools for treatment optimization can enable an efficient personalized approach. This can support the process where advanced, and often very costly, treatments are provided to patients who are predicted to benefit the most from them. The approach has potential to impact treatment optimization in large patient groups.
AI-driven analysis and knowledge transfer between multiplex and brightfield microscopy
Natasa Sladoje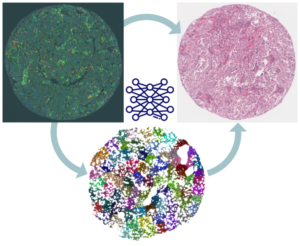
Uppsala University
Multiplex in situ immunofluorescence staining and imaging (multiplexIF) allows spatial quantification of the cellular repertoire of patients’ cancer tissue. MultiplexIF methods combined with AI-analysis are expected to reveal the mechanisms behind individual responses to immuno-therapy and to significantly advance cancer research. However, multiplexIF is a complex, labor-intensive and expensive technology, and its wider clinical implementation is unrealistic. To maximize clinical relevance of multiplexIF-based information, it is imperative to bring it closer to clinical practice – to reduce the reliance on expensive and exclusive imaging techniques, but also to enable pathologists to position the findings in their frame of experience. Approaches that rely on hematoxylin and eosin (HE) stainings as a diagnostic standard are therefore still being developed, despite the much lower information content of this modality.
Our team aims to develop AI-based approaches to transfer the predictive power of multiplex image information to HE images. We will thereby maximize clinical applicability and utilization of gained knowledge on cancer immuno-therapy, providing reliable and widely accessible information and analysis tools at every pathology lab.
Rapid 4D MR Flow Imaging
Petter Dyverfelt
Linköping University
Time-resolved, three-dimensional magnetic resonance flow imaging (4D Flow MRI) permits comprehensive evaluation of cardiovascular blood flow. The transition into the clinic is hampered by long data acquisition and data reconstruction times. Image reconstruction based on deep learning has the potential to offer a new paradigm for rapid reconstruction of fast, heavily undersampled MR data acquisitions. The purpose of this proposal is therefore to explore rapid 4D Flow MRI image reconstruction based on deep learning. We will compare two different approaches, one approach based on a U-Net architecture and one approach based on a residual network (ResNet) architecture. Successful accomplishment of this project will lead to reduced data acquisition times of 4D Flow MRI, which will reduce patient discomfort. Additionally, the rapid reconstruction methods developed in this project has the potential to permit 4D Flow MRI data acquisition and data reconstruction times of less than one minute. This would open up for broad clinical use of 4D Flow MRI.
ST-AI-PROCAP: Spatial Transcriptomics, AI & PROCAP
Jens Sörensen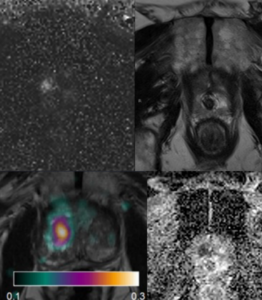
Uppsala University
Currently used MRI techniques for prostate cancer do not provide accurate estimates of tumor aggressiveness, which is an important factor for prognosis and treatment choice. Prostate cancer perfusion, or tumor blood flow, measured with H2O-positron emission tomography (H2O-PET), on the other hand, has been found to be an effective tool for assessing degree of aggressiveness. H2O-PET is however not widely available. In this project, we will evaluate the performance of different MRI perfusion techniques for prediction of prostate cancer aggressiveness in comparison with H2O-PET and with histopathological evaluation (Gleason score) as reference. We will further use a novel technique for spatial localization of different RNA strands in histopathology slides (spatial transcriptomics), which will be spatially compared with PET and MRI. This will enable identification of key proteins influencing cancer aggressiveness, and patterns of PET and MRI findings that relate to cancer aggressiveness. AI techniques will be used for prediction of malignant features from histopathology, and for assessing the relation between tumor bloodflow and gene expression.
Precision microcirculation imaging for decision support in patients with lower-limb ischemia
Ingemar Fredriksson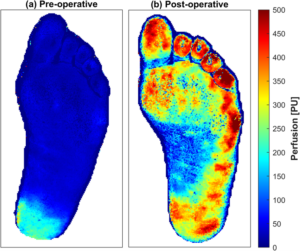
Linköping University
Impaired vessel function in the lower limb leads to ischemia causing pain, wounds, necrosis and ultimately amputations. The problem can originate from larger arteries, the microvasculature, or a combination of both. However, current clinical practice only evaluates the macrocirculation, while the microcirculation is left unattended. In the first phase, this project will merge the analysis of multi-spectral and multi-exposure laser speckle contrast images to assess the microcircular status. Applying the neural network on each multi-channel pixel in measured images will lead to precision imaging of speed-resolved perfusion and oxygen saturation at video rates. This has the potential to be a new biomarker for lower-limb peripheral tissue status. In the second phase of the project, such images will be used to develop a convolutional neural network-based prediction model for wound healing potential. If implemented in clinical practice, such a tool would lead to faster and more accurate treatment decisions, saving healthcare resources as well as patient suffering and limbs.
Automated identification of vertebral fractures in VFA images with deep learning
Mattias Lorentzon
Göteborg University
Vertebral fractures (VFs) are the most common osteoporotic fracture and among the strongest predictors for future fracture. VFs can be identified by vertebral fracture assessment (VFA) from lateral spine imaging by dual-energy X-ray absorptiometry (DXA). Skilled physicians with expertise in diagnosing VFs by VFA are lacking in Sweden. Thus, there is a substantial need for automated methods for VF identification. We will develop a deep learning (DL) model for automated VF diagnosis able to (1) detect VFs, (2) classify the severity of the VFs, and (3) predict incident fractures from DXA images, independently from clinical risk factors. The DL model will use ensemble convolutional and/or vision transformer networks. A DL method that can diagnose VFs on par with expert readers, could improve the identification of both symptomatic and asymptomatic VFs, leading to earlier diagnosis of osteoporosis and therapeutic intervention resulting in reduced number of fractures in the older population.
Pediatric brain tumor diagnostics using AI-based digital pathology
Neda Haj-Hosseini
Linköping University
Histology slides, when digitized, can be shared with a wider society and automatically analyzed. AI provides a powerful tool for analysis of these images on a large scale and enables a fully automatic process, in addition to adding accuracy and consistency to the analysis. There are however several challenges to be addressed in analyzing the images including the large size of the data, categorization of complicated features, generalization of the algorithms to different datasets and the explainability of the developed methods. In this project we aim to implement AI-methods on pediatric brain tumor histology images, as a decision support in the diagnostic procedure of the pediatric brain tumors and address a some of the mentioned challenges.
AI-based multiplex tissue analysis to advance diagnostics for cancer patients in the context of immunotherapy
Patrick Micke
Uppsala University
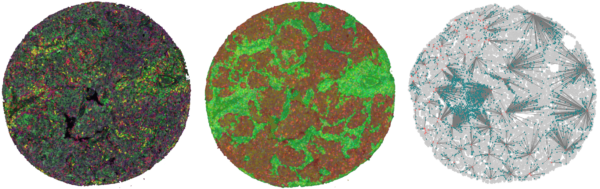 Cancer immunity is based on the interaction of a multitude of cells in the spatial context of the surrounding tissue. Because of its complexity, it is hitherto difficult to use this tissue information to help clinical decision-making for cancer immunotherapy. To address this shortness, we have developed a multiplex-multispectral pipeline that provides images of cells and structures in tissues from cancer patients that go beyond traditional diagnostics. To exploit the full potential of these multi-layer images, we will develop state-of-the-art deep learning models for predicting patients’ survival and response to immunotherapy, and combine them with the explainable artificial intelligence (XAI) tools. This will ensure their interpretability by pathologists and facilitates an early implementation into clinical diagnostics. We hope to clarify the biological mechanism of cancer immunity and explain why patients are responsive or resistant to immune-modulating agents. This will hopefully lead directly to novel prognostic and predictive markers for clinical use that can guide immunotherapy for lung cancer patients.
Cancer immunity is based on the interaction of a multitude of cells in the spatial context of the surrounding tissue. Because of its complexity, it is hitherto difficult to use this tissue information to help clinical decision-making for cancer immunotherapy. To address this shortness, we have developed a multiplex-multispectral pipeline that provides images of cells and structures in tissues from cancer patients that go beyond traditional diagnostics. To exploit the full potential of these multi-layer images, we will develop state-of-the-art deep learning models for predicting patients’ survival and response to immunotherapy, and combine them with the explainable artificial intelligence (XAI) tools. This will ensure their interpretability by pathologists and facilitates an early implementation into clinical diagnostics. We hope to clarify the biological mechanism of cancer immunity and explain why patients are responsive or resistant to immune-modulating agents. This will hopefully lead directly to novel prognostic and predictive markers for clinical use that can guide immunotherapy for lung cancer patients.
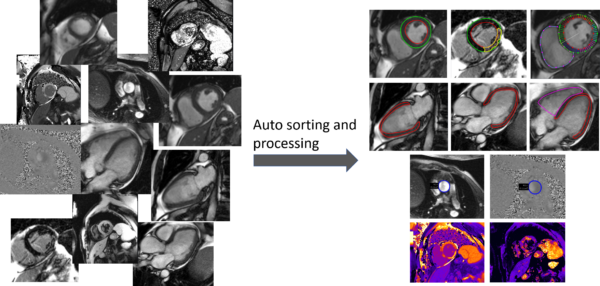
Taming the clinical wilderness
Jane Tufvesson
Region Skåne
AI tools hold a promise to increase productivity and quality in healthcare. However, many promising AI-tools never reach a large-scale adoption in the clinical routine, likely because the systems are typically trained and tested in well-behaved environments but fail in the clinical wilderness where the image data is not well structured. The purpose of this project is to develop machine learning based tools that can tame the clinical wilderness by being able to robustly identify, load, and process unstructured image data. We will accomplish this by using a machine learning approach for image classification based on both image content and image meta data. Specifically, we will focus on processing cardiac magnetic resonance images where one exam often consists of >100 image series and 10 000 images. The project is of high clinical importance by being a prerequisite for enabling automation of image analysis task in cardiac MRI using AI-tools. Image analysis automation saves valuable resources and greatly increases clinical productivity and reproducibility. Besides the clinical implications, a fundamental impact of the project is to enable research projects that otherwise would not be feasible due to shear amount of manual work involved.
Diagnosis and Subclassification of Non-Pigmented Skin Cancers Using Machine Learning Algorithms
Ida Arvidsson
Lund University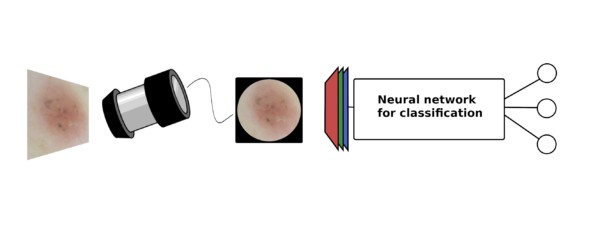 Skin cancer is the most common cancer worldwide. In Sweden, incidence of all skin cancer has increased dramatically the last decades and continue to increase at a fast pace of approximately 5-6% per year. To minimize associated morbidity and mortality of skin cancer, early detection is of substantial importance. However, in early developmental stages it can be difficult to differentiate skin cancer from benign lesions. Most studies on AI detected skin cancer have focused on pigmented tumours and more specifically the detection of malignant melanoma. Much less resources are guided to the less deadly, but more abundant and thereby quite costly, non-pigmented skin cancers. Classifying non-pigmented skin cancer is also considered a harder problem than its pigmented counterpart. This project aims to construct a machine learning algorithm for differentiation between superficial, nodular, and invasive basal cell carcinoma. Most studies use transfer learning, which is an efficient technique when the amount of data available is limited. We plan to further develop the idea, by for example utilizing contrastive learning or variational autoencoders, allowing the usage of unlabelled data together with labelled data.
Skin cancer is the most common cancer worldwide. In Sweden, incidence of all skin cancer has increased dramatically the last decades and continue to increase at a fast pace of approximately 5-6% per year. To minimize associated morbidity and mortality of skin cancer, early detection is of substantial importance. However, in early developmental stages it can be difficult to differentiate skin cancer from benign lesions. Most studies on AI detected skin cancer have focused on pigmented tumours and more specifically the detection of malignant melanoma. Much less resources are guided to the less deadly, but more abundant and thereby quite costly, non-pigmented skin cancers. Classifying non-pigmented skin cancer is also considered a harder problem than its pigmented counterpart. This project aims to construct a machine learning algorithm for differentiation between superficial, nodular, and invasive basal cell carcinoma. Most studies use transfer learning, which is an efficient technique when the amount of data available is limited. We plan to further develop the idea, by for example utilizing contrastive learning or variational autoencoders, allowing the usage of unlabelled data together with labelled data.
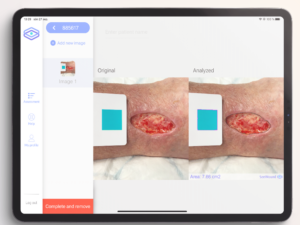
An AI-based clinical decision support device for bedside wound care of difficult to heal wounds
Folke Sjöberg
Linköping University
Hard-to-heal wounds are a growing societal problem that creates great suffering and costs Swedish health care 10-20 billion SEK per year (2-4% of the healthcare budget). Patients with hard-to-heal wounds are often treated by nurses and assistant nurses in primary care and home care settings, a group of professionals with limited training in advanced wound care and without any efficient tools or aids to enable them to provide the optimal treatment. The aim of this project is to address this challenge by improving the application SeeWound currently used to monitor wound healing and to further develop the underlying AI algorithms to also identify other aspects central to wound healing. The resulting system will enable healthcare professionals to offer patients with hard-to-heal wounds more continuous and efficient care across the full healthcare provider continuum (from specialist clinics to primary care and municipality care) with the goal of limiting the suffering and shortening the healing times for patients with these types of wounds.
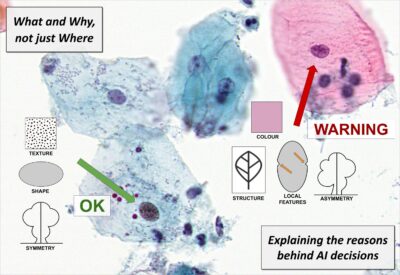
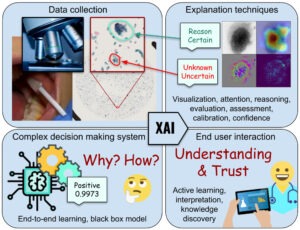
What and Why, not just Where – More informative explanations of deep learning-based cancer diagnostics
Joakim Lindblad
Uppsala University
With the growing prevalence of AI-techniques to support decision making throughout society, there is an urgent demand to explain how these decisions are made. This has led to the creation of the field of Explainable AI (XAI). Several techniques have been developed to illuminate the process; however, the vast majority of methods offer explanations of the type where in the data the important factors are appearing. These saliency, activation, or attention methods highlight where the network is “looking” when reaching a decision, but do not answer what the network finds as important at these positions. Is it colour, shape, texture? Neither do they provide information on how a combination of properties leads to the network output. Is it that the observed cell is unusually large, in combination with having a coarse texture, which leads to it being classified as malignant, or is it the combination of a thick nuclear envelope and the particular hue of the cytoplasm? This project aims to develop methods which can deliver such information – essential for confident usage of AI for critical tasks such as medical diagnostics, but also for gaining improved insight in the underlying phenomena. We explicitly target the creation of practically useful XAI methods for trustworthy and interpretable cancer detection and diagnostic support.
Self-supervised learning in coronary CTA
Ola Hjelmgren
Sahlgrenska University Hospital
The aim of this project is to develop a deep learning model for coronary computed tomography angiography (CCTA) using techniques from the field of self-supervised learning and uncertainty quantification. To date, we have successfully developed a fully supervised deep transformer-based model for automatic segmentation of coronary artery vessels and coronary plaques in CCTA. In this project, we will improve this model by 1/ boost the model performance with self-supervised pre-training on 4000 unlabeled CCTA datasets, 2/ enable model interpretability by incorporating confidence quantification, and 3/ evaluate on 1000 unlabeled images and compare the results to previously collected tabular (image-level) data. In order to so, we have a large annotated dataset (n>600) and a very large cohort for self-supervised learning (n>5000), as well as a research team including a senior radiologist expert, a deep-learning expert and one full-time engineer.
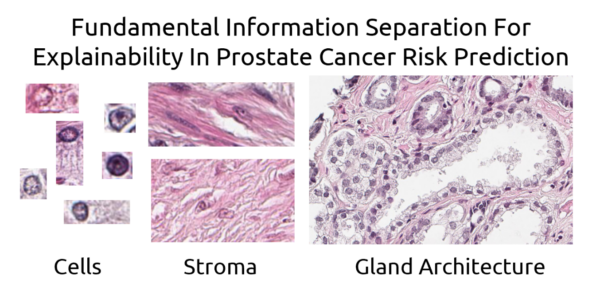
What is the basis of our AI based prostate grading index?
Anders Drotte
Spearpoint Analytics AB
The established method to grade the malignancy of prostate cancer, Gleason grading, is only based on the glandular architecture of the prostate tissue. We have trained an AI to grade the cancer by showing it tissue images and giving it the associated patient outcome. We believe that the resulting AI based malignancy index is based not only on glandular architecture but also on nuclear morphology and changes in the structure of the stroma. In this project we will use ablation techniques and similar approaches to explore what influence the different aspects of the tissue has on the resulting prognostic index. The goal is to reach a better understanding of what determines the outcome for the patient and what is the best way of predicting the outcome in an understandable and reliable way. A reproducible and reliable way of grading the prostate cancer is important in planning the treatment avoiding over- and under- treatment of the patients.
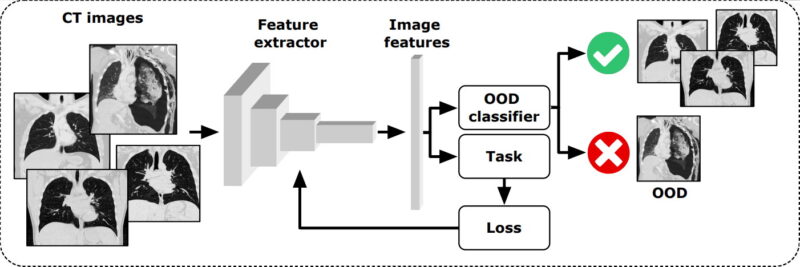
Deep learning-based out-of-distribution detection of computed tomography images
Fredrik Kahl
Chalmers University
There exists a plethora of deep learning (DL) methods to analyze computed tomography (CT) images, aimed at diagnostics, prognostics, segmentation etc. However, there are always abnormal (a.k.a. out-of-distribution, OOD) cases, that are not well represented in the training data, such as patients with artificial implants, atypical anatomy, uncommon BMI, etc. Application of most DL models on such cases can thus produce irregular results which are highly unreliable with large errors that may not easily be noticed.
We will here develop an unsupervised convolutional neural network-based model to analyze CT images and detect OOD images. Our model will flag OOD cases and indicate whether these cases are unreliable for use as training or testing with other trained DL models, to prevent spurious results. To classify OOD cases, we will investigate the distribution of the embedded space using different distance metrics, implement an autoencoder reconstruction model, and one self-supervised multi-class model based on geometric transformations.
To safely and reliably deploy DL models in real world clinical applications with high performance, it is crucial to detect irregular inputs that deviate from the models known capability, making OOD detection paramount for robust and dependable clinical models.
Advanced Body Composition Analysis in SCAPIS using Deep Learning and Image Registration
Joel Kullberg
Uppsala University
This project aims to extend the currently planned analysis of body composition from the 3-slice CT data collected in SCAPIS with three main contributions.
- Use deep learning to improve segmentation accuracy and to extend the analysis to more detailed depots and measurements.
- To utilize previous technology from our research group to develop an inter-subject image registration pipeline for the SCAPIS data. This will allow voxel-wise analysis of associations between the CT images (tissue volume from Jacobian determinants and Hounsfield units) with all other biomarkers collected in SCAPIS
- Tailor our previously developed pipeline for deep regression of MRI data to these 3-slice CT scans for deep learning-based prediction of relevant phenotype and disease information. The use of saliency analysis in combination with image registration furthermore allows for aggregated/cohort saliency analysis enabling biological interpretation of the predictions.
The project aims at improving the use of the 3-slice CT image data collected. This will likely improve currently planned research studies and also open up for new types of studies. It thereby has the potential to benefit patients affected by all diseases studied in SCAPIS, including metabolic, cardiovascular, and chronic obstructive pulmonary disease.
Breast Cancer Diagnosis using Pocket-sized Ultrasound Device and Deep Learning
Ida Arvidsson
Lund University
Breast cancer is the most common form of cancer among women. While screening works relatively well for early detection in high-income countries, there is no corresponding solution in low- and middle-income countries. It does not seem likely that the breast cancer care can be copied from high-income countries, due to its high cost and the lack of highly developed health care infrastructure. Also, the availability of diagnostic procedures, i.e., mammography facilities, trained radiographers and radiologist are often lacking. Therefore, we suggest the development of an accessible breast diagnostic tool, consisting of a technically enhanced low-cost pocked-sized ultrasound device paired with a smartphone-based artificial intelligence algorithm.
The goal of this project is to develop such an algorithm with existing data and tune the algorithm to work well when the pocket ultrasound probe is used. Initially, supervised training and validation of a convolutional neural network will be performed, with the aim to achieve a network which generalizes well. The network will be evaluated on images obtained from the pocket ultrasound and the images will in the end be presented and analysed on a smartphone.
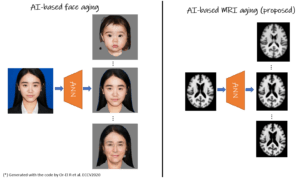
Synthetic MRI aging
Rodrigo Moreno
Royal College of Technology
One challenge in image analysis of Alzheimer’s patients is to disentangle the effects of aging from the disease itself. For this, it is necessary to use a large number of images of healthy subjects at every specific age, which is not available in the current longitudinal imaging datasets. In this project, we will develop an AI-based solution to generate magnetic resonance images (MRI) at different ages by simulating the changes in the images due to aging. In particular, given a structural MRI acquired at a specific age, the solution will be able to generate MRI of the same subject at older and/or younger ages. AI-based diffeomorphic registration will be used extensively in this project since, unlike standard generative models, it is able to preserve the anatomy of the brain.
We will validate the method for mild cognitive impairment (MCI) by comparing the subjects’ actual time course with the ones of simulated healthy and MCI subjects. Having a tool to accurately generate new time points of MRI data will have a big impact on the diagnostic work-up by helping to predict clinical progression of patients over time (prospective), or providing information about premorbid states (retrospective).
COVID-19 Survivors Assessed with Diffusion MRI and Artificial Intelligence
Evren Özarslan
Linköping University
COVID-19 infection is a new disease posing numerous clinical questions that will be investigated by the research community in the coming years. Of interest to us are the neurological symptoms experienced by some survivors, which have a significant impact on their quality of life.
In this project, we intend to employ state-of-the-art MRI methods sensitized to random movements of water molecules for assessing microstructural alterations associated with the infection and/or its treatment. To this end, we employ a new microstructure-sensitive diffusion MRI technique, which has not been fully incorporated into clinical practice. Our goal is to obtain more detailed information on the tissue structure and hopefully elucidate some of the pathogenesis of COVID-19 on the central nervous system. In this endeavor, artificial intelligence will play an important role by enabling the visualization of the intravoxel tissue composition probed by diffusion MRI.
The development of the data analysis framework including the visualization of the diffusion MRI data will facilitate the translation of the technique into clinical use, making it available for studying other clinical challenges.
Multimodal imaging and information fusion for confident image-based cancer diagnostics
Nataša Sladoje
Uppsala University
Cancer is a complex disease; its different causes and types strongly affect patient treatment and prognosis. Through exploring and developing novel techniques for multimodal information fusion we aim to improve understanding of the disease, its causes and progression, and enable reliable early detection and confident differentiated diagnosis, thereby providing a solid basis for treatment planning. In this collaboration between Uppsala University and Center for Clinical Research Dalarna we will, through powerful AI-based data fusion, combine information from a range of imaging techniques to capture complementary information about a specimen. We hypothesize that fusion of heterogeneous information about the specimen will be beneficial in a number of ways, enabling (i) improved and differentiated ground truth data for learning and evaluation, where additional modalities support the cytopathologist towards more reliable annotation; (ii) improved performance of our existing AI-based cancer diagnostics decision support system, through (direct or indirect) use of relevant information from additional modalities; (iii) increased explainability, through ability of the system to indicate and correlate active virus infections with the cancer; (iv) improved understanding of the disease and its causes, enabling improved patient treatment (personalized medicine).
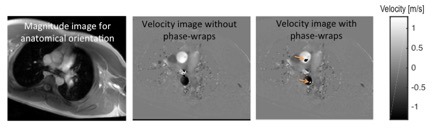
Expanding the Clinical Applicability of MR Flow Measurements using Deep Learning
Petter Dyverfelt
Linköping University
Magnetic resonance (MR) flow measurements are used clinically for determination of cardiac output, shunt flow as well as aortic and pulmonary regurgitation. However, since the introduction of the technique about three decades ago, MR flow measurements have been hampered by image artifacts which compromise the clinical applicability and reliability of the technique. Although the artifacts are well-known, current methods used to suppress or correct the artifacts only provide a partial solution. The purpose of this project is to develop and evaluate deep learning techniques for correction of known artifacts in MR flow measurements. Successful accomplishment of this project will reduce the number of re-acquisitions that need to be made when MR flow measurements are affected by phase wraps, and will also improve the reliability of the results. By aiming to provide direct solutions to factors that negatively impact and sometimes hinders currently used clinical methods, the project has the potential to reach immediate clinical usefulness.
Automatic software using convolutional neural networks for the segmentation of coronary vessels in coronary CT
Ola Hjelmgren
Gothenburg University
The aim of this project is to develop a deep learning model for the segmentation of coronary arteries in coronary computed tomography angiography (CCTA) images. This model will later be used to develop software for the automatic segmentation of coronary artery plaques. Automatic detection and segmentation of coronary plaques is a promising tool, both for individual risk assessment of previously healthy individuals, but also as a promising tool for management and work up of clinically patient presenting with symptoms of acute coronary syndrome. With the help of effective decisions support software, assessment of CCTA could be made available to larger patient groups, in smaller hospitals and outside normal work hours. In patients with chest pain, CCTA can be used as a non-invasive tool to exclude coronary artery disease or, if present, to analyze plaque burden and composition. These data can be used to identify patients with high risk of developing myocardial infarction and can be used in improving prevention.
AI-tool for emphysema assessment in SCAPIS CT images
Mats Lidén
Region Örebro Län
Chronic obstructive pulmonary disease (COPD) is a common and progressive lung disease generally caused by smoking. Emphysema, the irreversible destruction of the lung tissue, recognizable in computed tomography imaging (CT), is one key feature of COPD.
In the national Swedish CArdioPulmonary bioImage Study (SCAPIS), 30,000 Swedish men and women aged 50 to 64 years have been investigated with detailed imaging and functional analyses including a thoracic CT scan. One of the aims in SCAPIS is to predict COPD and provide better understanding of the disease. Quantitative assessment of emphysema is essential for using imaging in COPD research. For SCAPIS research purposes, the emphysema quantification needs to be reproducible, consistent with visual assessment, fully automatic and perform well in the specific cohort.
In this innovation project, the objective is to create an automated method, specifically developed for SCAPIS image data, for assessing the emphysema in CT images, using annotations provided by multiple readers in a previous innovation project. An automated method permits the inclusion of quantitative emphysema image data in future studies in the SCAPIS cohort. With increased knowledge, quantitative imaging may also be beneficial for patients in routine clinical COPD care.
Segmentation of Scoliotic Backs Using Artificial Intelligence – Application for 3D Printing
Helen Fransson
Medviso AB
Using a 3D printed model of the pre-operative spine allows the surgeon to better plan the challenging surgery. Better planned surgeries allow for safer surgeries and reduced surgery time. 3D printing is today used more and more in the daily work in hospitals to optimize patient care. However, what is needed for a more wide-spread clinical usage is faster and better 3D segmentation tools, and better support and start-up guidance for hospitals how to implement the technique in clinical routine. This project aims to meet both of these needs.
In the project we will implement automated spine segmentation algorithms based on machine learning. The algorithms will be developed together with surgeons and integrated into existing CE-marked software available for clinical use. We will incorporate this algorithm in a product consisting of software and a support package for hospitals around the world on how to implement the technique in clinical routine.
The developed technique has a great potential to improve patient care by making 3D printing more accessible for pre-surgical planning and thus shorten operation times. It has been shown that shortened operation times translates to reduced risk for infections and complications for the patient.
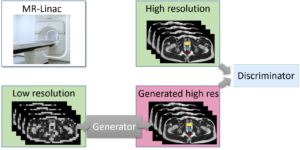
Image processing in MRI guided real-time adaptive radiotherapy
Robin Strand
Uppsala University
As one of the first two hospitals in Scandinavia and among the early adopters globally, the MR-Linac has been installed at the Uppsala University Hospital. The MR-Linac combines MR and radiation therapy and offers the possibility to adapt the radiation to the moving target in real-time during radiotherapy treatment.
The internal geometry of the patient can change significantly due to breathing, peristaltic, or just simply because the patient relaxes or get tense. Today, this is handled by adding geometrical margins. The obvious drawback is irradiation of healthy tissue.
This project aims at developing methods to tailor the radiation online, based on the stream of images coming from the MR during the radiation, i.e. real-time adaptive radiotherapy (RT-ART).
In order to achieve this aim, this project will develop methods for real-time segmentation of target and organs at risk in pelvic treatments. The obtained higher precision and certainty can be utilized to deliver the correct dose to the target while minimizing the healthy tissue complications.
Efficient and automatic tools are key to ensure that as many patients as possible can benefit from these advanced treatment methods. One of the first patient groups treated with the MR-Linac is palliative prostate patients. More patient groups will follow.
Multimodal Algorithm in Traumatic Brain Injury Prognostics
Jennifer Ståhle
Karolinska University Hospital
Traumatic Brain Injury (TBI) is the leading cause of death and disability in people <40yrs worldwide. Standard imaging at admission is non-contrast CT due to its wide availability and low acquisition time. The initial TBI results from the mechanical forces of impact leading to focal or diffuse brain injury patterns. Following the days after trauma a complex cascade of intracranial events can result in secondary brain injury with further brain damage.
Immediate and accurate interpretation of the CT images is crucial to assess the need for neurosurgical treatment or predict potential secondary injuries. While initial assessment is performed in settings ranging from Level 1 Trauma centers to more remote ER locations, interpretation has considerable inter-observer variability. Development of Machine Learning algorithms provide crucial steps in reducing error and refining diagnosis in the trauma setting, leading to improved outcome for patients suffering from TBI. Further on multimodal algorithms has the potential to improve prediction of secondary injuries, important steps toward future customized treatment.
With experience from neuroradiology, neurosurgery and computer science the project aims to develop a diagnostic and prognostic AI tool by integrating CT imaging data with clinical patient data. If successful, our results can contribute to develop future diagnostic and decision support tools to be tested in a clinical setting.
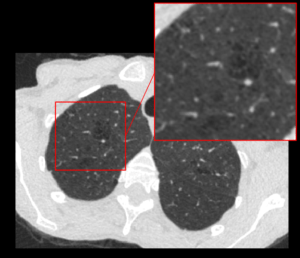
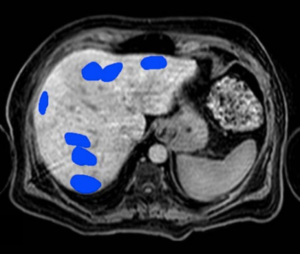
Biopsy Image Based AI Prostate Grading, Trained Directly on Patient Outcome
Anders Drotte
Peaky Networks AB
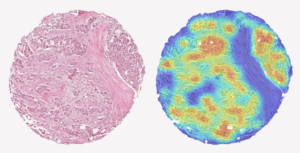
A tissue sample from prostate cancer and a AI produced heat-map showing the aggressiveness of the cancer.
Today prostate cancer is analyzed by a pathologist. The currently used Gleason Grading is known to have poor reproducibility and only moderate correlation to actual patient outcome. This problem will remain when an AI is trained to reproduce the Gleason gradings.
Our approach is instead to train the AI system directly on patient outcome. We have achieved promising results from a system trained on a large set of tissue samples from patients with known outcome. In this project we will further develop and extend the analysis with tissue samples collected with another method.
The samples will be collected from an ongoing study at Akademiska sjukhuset in Uppsala in which we will use biopsies from patients that are followed until final outcome is known.
The goal is to create a score that will better help the urologist to determine the best treatment for the individual patient. The effect will be avoiding unnecessary overtreatment made today without increasing the risk of unfavorable, in worst case fatal, outcome.
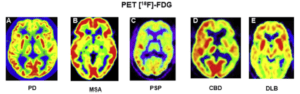
Evaluation of deep learning to predict the diagnosis of LBD using 18F-FDG PET
Kobra Etminani, PhD
Halmstad University
The main purpose of this project is to compare deep machine learning methods to human visual interpretation in clinical applications. We want to evaluate whether Artificial Intelligence (AI) models (including shallow and deep learning algorithms) could be trained to predict the final clinical diagnoses in patients who underwent 18F-FDG PET scans of the brain and, once trained, how these algorithms compare with the current standard clinical reading methods in differentiation of patients with final diagnosis of LBD or no evidence of dementia. We hypothesized that the AI model could detect features or patterns that are not evident on standard clinical review of images (both visual and quantitatively with the available commercial programs for brain quantification) and thereby an earlier detection of pathology, improving the final diagnostic classification of individuals.
There are various problems within this domain including intra-observer differences and limited number of nuclear medicine specialists with experience in 18F-FDG PET brain scans. We believe that we can contribute to develop an AI algorithm that is more invariant to different nuclear medicine specialist and help in coming faster to a diagnosis from the images thus improving healthcare for these patients.
Trustworthy AI-based decision support in cancer diagnostics
Joakim Lindblad
Uppsala University
To reach successful implementation of AI-based decision support in healthcare it is of highest priority to enhance trust in the system outputs. One reason for lack of trust is the lack of interpretability of the complex non-linear decision-making process. A way to build trust is thus to improve humans’ understanding of the process, which drives research within the field of Explainable AI. Another reason for reduced trust is the typically poor handling of new and unseen data of today’s AI-systems. An important path toward increased trust is, therefore, to enable AI systems to assess their own hesitation. Understanding what a model “knows” and what it “does not know” is a critical part of a machine learning system. For a successful implementation of AI in healthcare and life sciences, it is imperative to acknowledge the need for cooperation of human experts and AI-based decision-making systems: Deep learning methods, and AI systems, should not replace, but rather augment clinicians and researchers.
This project aims to facilitate understandable, reliable and trustworthy utilization of AI in healthcare, empowering the human medical professionals to interpret and interact with the AI-based decision support system.
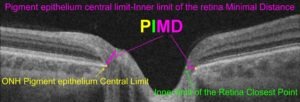
Segmentation of the waist of the nerve fiber layer by AI for clinical evaluation of progression of glaucoma
Per Söderberg
Uppsala University
Glaucoma is a major cause of vision loss. Objective evaluation of glaucoma with visual field testing is insufficient for early detection and early establishment of progress of the disease. In the optic nerve head, nerve fibers conducting the visual information from the eye to the brain merge into the optic nerve. Optical coherence tomography images the structure of the optic nerve head with close to micrometer resolution. We have developed a semi-automatic strategy that allows angularly resolved measurement of the waist of the nerve fibers at the optic nerve head, PIMD(angle). We have proven the feasibility of the strategy, and that PIMD(angle) measurement provides; much better resolution, can be obtained much quicker and with a lot less strain on the patient, than visual field testing.
Now, development of a fully automatic software is required for implementation in routine clinical work. We have demonstrated that it is feasible to use artificial intelligence (AI) for fully automatic detection of PIMD(angle). In this project a fully automatic method will be; developed, evaluated against a large dataset of semi-automatically classified volumes of optic nerve heads in glaucoma patients, and applied to new measurements of the optic nerve head in no pathological humans.
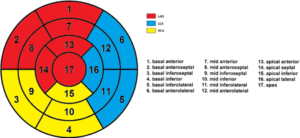
AI-based evaluation of coronary artery disease using myocardial perfusion scintigraphy based on deep learning
Miguel Ochoa Figueroa, PhD
Region Östergötland
The project aims to develop an AI algorithm for the evaluation of coronary artery disease using myocardial perfusion scintigraphy (MPS) based on deep learning convolutional neural networks. Analysis of perfusion acquisitions will be performed based on the American Heart Association’s (AHA) standard 17-segment model (Figure 1). In order to create a robust AI algorithm the patient selection will be very exclusive and made by only including those patients who have gone through invasive coronary angiography (ICA). The AI algorithm will also include patients with low or very low probability for ischemia according to the pre-test probability by Gibbons et al [1].We will include around 750 patients which will fulfill the above stated criteria. Finally we will add some heart conditions, which present with typical patterns in the MPS such as left bundle branch block to also train the AI algorithm with these patterns.
The development of the AI algorithm will reduce intra-observer differences between doctors. Another problem is the limited number of nuclear medicine specialist with experience in MPS, which is also decreasing in the near foreseeable future. An automatic or even semi-automatic system will drastically improve the productivity and thus also improving health care.
Radiological Crowdsourcing for Annotation of Emphysema in SCAPIS CT Images
Mats Lidén, PhD
Örebro University
Chronic obstructive pulmonary disease (COPD) is a common and progressive lung disease generally caused by smoking. Emphysema, an irreversible destruction of the lung tissue, which can be assessed by CT imaging, is one key feature of COPD.
In the national Swedish CArdioPulmonary bioImage Study (SCAPIS), 30.000 Swedish men and women aged 50 to 64 years have been investigated with detailed imaging and functional analyses including a thoracic CT scan. One of the aims in SCAPIS is to predict COPD and provide better understanding of the disease. Automated emphysema quantification in the acquired CT images is, therefore, expected to be effective for future research and may emphasize the clinical role of imaging in COPD.
The purpose of the present AI project is to develop a method for quantifying the emphysema extent in CT images acquired in the SCAPIS Pilot project in Gothenburg 2012. This includes evaluation of an alternative approach for obtaining radiological annotations – radiological crowdsourcing. Instead of recruiting a small number of thoracic radiologists for the time consuming annotation task, the annotation is split into a very large number of small annotation tasks that can be performed by a large number of participating radiologists.
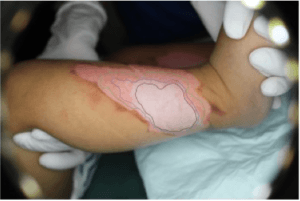
AI-based Decision Support System for Burn Patient Care
Tuan Pham, PhD
Linköping University
Burns are among the most life-threating of all injuries and a major crisis for the global public health with an implication to a considerable health-economic impact. In the European Union, burns are known as the most common fatal injuries after transport accidents, falls, and suicide.
Burns are classified into several types by depth: superficial dermal, deep dermal, and full thickness burns. The burn extent of a patient is quantified as the percentage of total body surface area (%TBSA) affected by partial thickness or full thickness burns. The initial assessment of %TBSA is crucial for the continued care.
To achieve precision burn assessment for optimal clinical decision making, our aims are to use digital color images and develop AI tools for 1) complex burn depth prediction, 2) measurement of body surface area, and 3) calculation of %TBSA in a precise, easy-to-use, and cost-effective way. The proposed AI-based decision support system will be extended for the management of chronic wounds, including diabetic ulcers, which our team members are planning to work on.
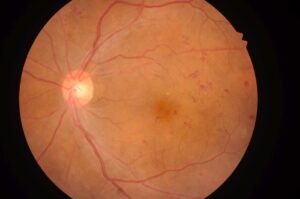
AI for a Healthy Eye
Christoffer Levandowski, PhD
QRTECH
The project will deliver an automated algorithm to make the analysis process of retinal images from diabetes patients more time-efficient, benefitting both health care professionals and patients.
The Swedish National Diabetes Register (NDR) covers 95 % of diabetes patients and had approximately 455 000 registered patients 2017. Diabetes patients are offered fundus screenings regularly, as a preventative measure. This results in large amounts of image data being collected and studied manually every year. The need for a more time-efficient alternative is evident. We propose a solution based on machine learning, enabled by the progress made in the field of artificial intelligence. By using machine learning methods, an automated algorithm can be created and used as a support system for diagnosis by the hospital staff. Instead of manually analysing the images, the algorithm will analyse them and provide filtered results as a foundation for diagnosis decisions. Thereby, the patients in need of treatment would be identified earlier, reducing distress and uncertainty. Furthermore, the reduction in tedious manual work would allow ophthalmic nurses usually responsible for analysing retinal images to redirect their focus to other tasks, such as treatment and patient contact.
Brain Tumor Segmentation Using Multi-Modal MRI
Anders Eklund, PhD
Linköping University
Brain tumors severely affect the quality of life for a large number of people. In this project the goal is to improve segmentation of brain tumors by combining novel diffusion imaging with deep learning. Existing approaches for segmenting brain tumors use 2D or 3D deep learning (convolutional neural networks), using different types of structural magnetic resonance imaging (MRI), for example T1 weighted images, T2 weighted images and T1 weighted images with gadolinium contrast agent.
In this project, we will take advantage of advanced diffusion imaging techniques featuring general gradient waveforms to further increase our information about the underlying micro structures. Using a multi-channel 3D CNN, the network will learn to combine the different MRI modalities to improve segmentation.
The project will lead to automatic segmentation and volume estimation of brain tumors, which could save time for neuroradiologists. This could be useful for comparing tumor size before and after treatment. The segmentation can also become more accurate, by combining several types of MR images, which can lead to a better foundation for treatment decisions.
Machine Learning for Reconstructive Surgery
Helen Fransson, PhD
Medviso
3D printing has the potential of revolutionising reconstructive surgery.Corrections of skeletal deformities are often very complicated and time consuming. Today most corrections of skeletal deformities are done on free hand thus depending much on the expertise of the surgeon and some deformities are so complicated that it is not possible to correct them.
In this project we will develop machine learning techniques for creating cutting/drilling guides and patient-specific implants for 3D printing. The techniques will be developed together with surgeons andintegrated into existing CE-marked software available for clinical use. The developed technique has a great potential to improve patient care by making it possible to correct very advanced deformities withhigh precision and lowered complication rates. Pre manufactured 3D printed implants will shorten operation times. It has been shown that shortened operation times translates to reduced risk for infections and complications, thus this can lead to better outcome for the patient and reduced costs.
Artificial Intelligence for Human Brain Structural Connectomics
Rodrigo Moreno, PhD
KTH Royal Institute of Technology
Connectomics aims at studying connectivity among different areas of the brain. Structural connectivity, which is the topic of this project, can be inferred by analyzing tractograms computed from diffusion MRI (dMRI). Structural connectomics is a promising tool for detecting connectivity impairments due to diseases. Unfortunately, the current state-of-the-art tractography methods have high sensitivity but very low specificity, which makes it difficult to perform structural connectivity analyses. Despite its potential, the current use of artificial intelligence (AI) in the field is relatively scarce, partially due to the orientation-dependent nature of dMRI data and the difficulty of having reliable ground truths.
In this project, we aim at developing an AI solution for structural connectivity analysis. These methods will be used to assess impairments in structural connectivity of patients with monoaural canal atresia (MCA), that is, patients that are born with one ear canal closed but have all the internal hearing organs intact. More specifically, the project include: a) development of a bundle-specific deep learning-based tractography method; b) test the method for performing structural connectivity analysis for MCA patients. Detecting the time point where early-stage impairments in brain connectivity appear in MCA patients is crucial to give them the best possible treatment.
Improving the Quality of Cardiovascular MRI Using Deep Learning
Tino Ebbers, PhD
Linköping University
In conventional MR Angiography, contrasts agents are crucial to obtain contrast between blood pool and surrounding tissue. However, the most common types of contrast agents are gadolinium based, which is contraindicated in patients with renal impairment. Moreover, recent studies have suggested that it can remain deposited in the brain after the examination. Phase-contrast angiography allows for the acquisition of angiographic images without the use of contrast agents, but the quality and spatial resolution of these data are not always sufficient.
The purpose of the proposed project is to develop and evaluate deep learning techniques to enhance image quality of cardiovascular MR imaging methods. The main focus of the work will be on improving important quality characteristics of cardiovascular MRI, such as blood-tissue contrast and image resolution, thus improving clinical applicability of these images for diagnostic purposes.
Successful implementation of these tools will result in less need for studies requiring contrast agents, contributing to a reduced risk for contrast-agent related complications and lower costs for clinical healthcare. Moreover, obtaining MR data with higher resolutions will improve the diagnostic quality of the images and facilitate shorter examination times, resulting in reduced patient discomfort and examination costs.
Image- and AI-Based Cytological Cancer Screening
Joakim Lindblad, PhD
Uppsala University
Oral cancer incidence is rapidly increasing worldwide, with over 450,000 new cases found each year. The most effective way of decreasing cancer mortality is early detection, which makes routine screening of patient risk groups highly desired. However, screening for oral cancer is not feasible with today’s methods that rely on painful tissue sampling and laborious manual examination by a medical expert. A consequence is that oral cancer is often discovered as late as when it has metastasized to another location. Prognosis at this stage of discovery is significantly worse than when it is caught in a localized oral area.
We will develop a system that uses artificial intelligence (AI) to automatically detect oral cancer in microscopy images of brush samples, which can quickly and without pain be routinely taken at ordinary dental clinics. We expect that the proposed approach will be crucial for introducing a screening program for oral cancer at dental clinics, in Sweden and the world. The project, which involves researchers from Uppsala University, Karolinska University Hospital, Folktandvården Stockholms län AB, and the Regional Cancer Center in Kerala, India, will greatly benefit from AIDA to turn developed methods into clinically useful tools.
Platform for Efficient Processing of Large Study Cohorts
Einar Heiberg, PhD
Medviso AB
New medical guidelines need large clinical studies to improve and change medical treatment and patient management. Currently, there is an unmet need for efficient tools that can analyze imaging data from large-scale study cohorts with 1,000-100,000 patients. Machine learning has a critical role in the analysis of large study cohorts, and may provide a paradigm shift in how support tools for clinical decision are developed.
Existing clinical analysis software packages are not adequate for analyzing large patient cohorts. The main reasons are 1) Workflow is not streamlined enough. 2) Manual interactions are required to load/save data and batch processing are lacking. 3) Existing tools do not save the results in an open format that allows re-processing to extract new data or for use with machine learning approaches.
Medviso has in close collaboration with Department of Clinical Physiology developed the software Segment for medical image analysis. This software platform is freely available for research purposes.
The purpose of his project is to improve the existing software platform with tools adopted to process large study cohorts to overcome all above obstacles.
AI Based Tumor Definition for Improved and Milder Radiation Therapy
Carl Sieversson
Spectronic Medical AB
The result of our development project will be used by physicians for planning of radiation therapy. In today’s radiotherapy, a physician manually delineates the exact contours of the tumor in a three-dimensional MR image. Based on this delineation, a set of intensity modulated radiation beams are directed towards the tumor, subjecting the tumor to the intended radiation dose while avoiding excessive radiation to sensitive surrounding healthy tissue. However, manually delineating a tumor is a time-consuming work characterized by a great deal of individual variation and uncertainty regarding the exact boundaries of the volume to be included. Recent scientific publications identify this arbitrariness as one of the largest sources of error in modern radiation therapy. The outcome of our development project will provide physicians with an automatically delineated target volume which can be used as a starting point for further manual refinements. These delineations will be generated by a deep-learning based software, which adaptively improves itself by studying how different physicians outlines different types of tumors.
Decision Support for Classification of Microscopy Images in Digital Pathology Using Deep Learning Applied to Gleason Grading
Anders Heyden, PhD
Lund University, Lund
Prostate cancer (PCa) is the second most common malignancy in men worldwide. In Sweden, the incidence is now close to 10 000 new cases per year. Correct identification of the stage and severity of the PCa on histological preparations can help the healthcare specialists predict the outcome of the patient and chose the best treatment options.
The best method of evaluating the severity of prostate cancer in tissue samples is based on the architectural assessment of a histologically prepared tissue specimen (from biopsy or surgery) by an experienced pathologist. The pathologist assigns a “Gleason grade” ranging from 1 (benign) to 5 (severe cancer). This type of cancer severity grading is highly correlated with prognostic outcomes and is the best biomarker in PCa to predict outcome. However, intra-observer differences between pathologists are a major problem. This results in costly repercussions; under-diagnosed patients may become more ill while over-diagnosed patients receive unnecessary treatment, in all decreasing the quality of life and increasing the costs for the healthcare system.
The project aims at developing decision support systems for Gleason scoring of microscopic images of prostate cancer based on deep convolutional neural networks (CNN). We will evaluate the results and obtain feedback on difficult cases from pathologists. The purpose of such a system is to obtain a more reliable estimate of the Gleason score and thus a correct treatment for the patient. The project is based at the Centre for Mathematical Society, Lund University, with close cooperation with the Department of Urology at Lund University Hospital and SECTRA in Linköping.
Interactive Visualization Tools for Verification and Improvement of Deep Neural Network Predictions
Ida-Maria Sintorn, PhD
Vironova AB, Stockholm
Understanding and being able to show what leads to a decision using AI and machine learning methods is important to gain acceptance for their use in clinical diagnostics. The purpose of this project is to adapt and develop interactive visualization tools to explain and test what regions and details in an image are important for the decision of the artificial neural network. Interacting with the visualizations and providing corrections and feedback will further train the neural network for improved performance. The interactive tools will, in addition, allow for convenient hypothesis testing regarding importance of image features by e.g. blocking regions or scales of images or part of a network and seeing the effect on the results.
Verifying what information decision support systems based their decision on is the key for rapid conversion of research results to clinical use. The tools to be developed in this project will hence, not be directly used clinically but rather support and serve as a catalyst for deploying other decision support systems in the clinics. The tools will be evaluated on clinical applications using electron microscopy for kidney diagnoses and light microscopy for cervical cancer screening.
Machine Learning for Automated Measurements of Liver Fat
Magnus Borga, PhD
AMRA AB, Linköping
Non-alcoholic fatty liver disease (NAFLD), a range of diseases characterized by steatosis, is associated with the metabolic syndrome and can lead to advanced fibrosis, cirrhosis, and hepatocellular carcinoma. Non-alcoholic steatohepatitis, a more serious form of NAFLD, is now the single most common cause of liver disease in developed countries and is associated with high mortality. Diagnosis and grading of hepatocellular fat in patients with NAFLD usually requires a liver biopsy and histology. However, as liver biopsy is an expensive, invasive, and painful procedure that is sensitive to sampling variability, the use of MRI as a non-invasive biomarker of liver fat has shown tremendous progress in recent years. Automation of this technology would further reduce costs for clinical use. Measuring fat in the liver is, however, not trivial since larger blood vessels and bile ducts need to be avoided in order to get accurate estimates of the liver fat fraction. Therefore, the aim of this project is to develop an automated method, based on machine learning, for placement of regions of interest (ROI) in which the liver fat can be quantified.
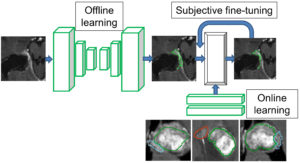
Interactive Deep Learning Segmentation for Decision Support in Neuroradiology
Robin Strand, Uppsala University
Many brain diseases can damage brain cells (nerve cells), which can lead to loss of nerve cells and, secondarily, loss of brain volume. Even slight loss of nerve cells can give severe neurological and cognitive symptoms. Technical imaging advancements allow detection and quantification of very small tissue volumes in magnetic resonance (MR) neuroimaging. Due to the enormous amount of information in a typical MR brain volume scan, and difficulties such as partial volume effects, noise, artefacts, etc., interactive tools for computer aided analysis are absolutely essential for this task.
In this project, we will develop and evaluate interactive deep learning segmentation methods for quantification and treatment response analysis in neuroimaging. Interaction speed will be obtained by dividing the segmentation procedure into an offline pre-segmentation step and an on-line interactive loop in which the user adds constraints until satisfactory result is obtained. See the conceptual illustration.
The successful outcome of this project will allow detailed correct diagnosis, as well as accurate and precise analysis of treatment response in neuroimaging, in particular in quantification of intracranial aneurysm remnants and brain tumors (Gliomas WHO grades III and IV) growth.
Automatic Detection of Lung Emboli in CTPA Examinations
Tobias Sjöblom, Uppsala University
Pulmonary embolism (PE) is a serious condition in which blood clots travel to, and occlude, the pulmonary arteries. To diagnose or exclude PE, radiologists perform CT pulmonary angiographies (CTPA). Each CTPA consists of hundreds of images. Manual CTPA interpretation is, therefore, not only time-consuming, but is also dependent on human factors, especially in the stressful conditions of emergency medical care. Several automatic PE detection systems have been developed, but none with acceptable accuracy for clinical usage.
Our team will combine expertise in clinical radiology, medical image processing and analysis, and diagnostic technologies to develop a system for fast, precise and reliable automatic identification of pulmonary embolization in CTPA examinations. The availability of a large set of annotated CTPA examinations, which is assembled by our team, is a particular asset which enables development and training of advanced deep learning methods to address the task. We therefore expect to reach performance required for usage in a daily clinical practice.
The resulting system will save precious time of both patients suspected of having PE and of expert radiologists during their daily clinical routines. This will, in turn, have a strong positive impact on the health system in Sweden, considering that CTPA is today one of the most common emergency CT examinations in the country.
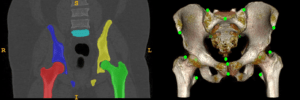
Simultaneous Landmark Detection and Organ Segmentation in Medical Images for Orthopedic Surgery Planning
Chunliang Wang, KTH, Stockholm
Landmark detection and organ segmentation are common tasks in medical image analysis. In this project, we aim to develop a multi-task deep neural network that can simultaneously detect multiple landmarks and segment several key structures. These multi-task networks can often deliver better results than the single-task models. The proposed algorithm will be tested in a hip surgery planning system using 3D CT images, where the segmentation results will provide graphical representation of the bones and the detected landmarks can be used to extract key measurements for the planning procedure. This will simplify and speedup the orthopedic surgery planning by replacing the most time-consuming step of landmark marking and joint attribute measurements in 3D CT images with automated tools. By simplifying the operation of the planning software, we hope to reduce risk-taking in clinical practice due to the cumbersome planning procedure and improve the outcome of the surgery.
AIDA MENY
KONTAKT
KALENDARIUM
MER INFORMATION
AIDA har sin fysiska bas på Centrum för medicinsk bildvetenskap och visualisering, CMIV, vid Linköpings universitet.CMIV har mångårig erfarenhet av att jobba med teknikutmaningar inom bildmedicin och att föra dess in i klinisk verksamhet. CMIV är också internationellt erkänt för sin kliniknära och tvärvetenskapliga spetsforskning inom bild- och funktionsmedicin, med verksamhet inom avbildning, bildanalys och visualisering. Du kan läsa mer om AIDA och CMIV på liu.se/forskning/aida